Welcome to the Center for Genomic Epidemiology
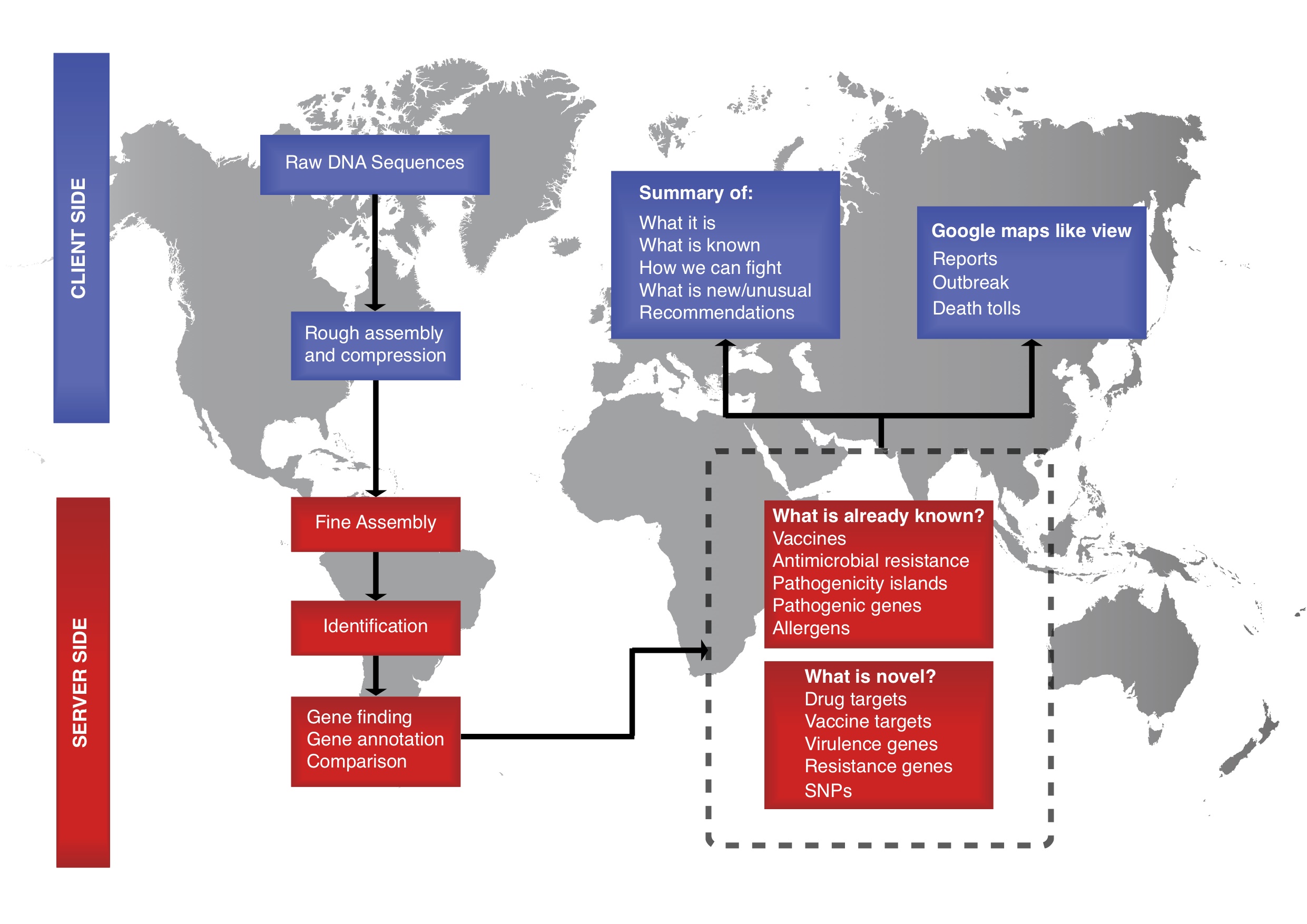
The use of sequencing technologies is currently transforming almost every aspect of biological science. In relation to infectious diseases, the advances are rapidly changing our scientific discoveries, as well as diagnostic and outbreak investigations. The ability to analyze sequencing data and take advantage of the rapid progress, is however, not equally distributed between institutions and countries.
The aim of the Center for Genomic Epidemiology (CGE) is to provide access to bioinformatics resources also for those with limited experience and thereby allow all countries, institutions and individuals to take advantage of the novel sequencing technologies. Doing so, we hope to facilitate more open data sharing around the world and provide more equal opportunities for all.
CGE is entirely non-commercial and operates a number of free online bioinformatics services. Funding is provided as core funding from the Technical University of Denmark (DTU) and from a range of public and private sources as listed below.If you want to read more about us and our research activities, please visit the Global Surveillance website.
News
Assembly-free typing of Nanopore and Illumina data through proximity scoring with KMA
September 2025Link to article....
Whole-genome prediction of bacterial pathogenic capacity on novel bacteria using protein language models, with PathogenFinder2
April 2025Link to article....
SpanSeq: similarity-based sequence data splitting method for improved development and assessment of deep learning projects
August 2024Link to article....
Scaling neighbor joining to one million taxa with dynamic and heuristic neighbor joining
January 2023Link to article....
SourceFinder: a Machine-Learning-Based Tool for Identification of Chromosomal, Plasmid, and Bacteriophage Sequences from Assemblies
November 2022Link to article....
ResFinder – an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes
January 2022Link to article....
MINTyper: an outbreak-detection method for accurate and rapid SNP typing of clonal clusters with noisy long reads
April 2021Link to article....
Automated download and clean-up of family specific databases for kmer-based virus identification
October 2020Link to article....
CRHP Finder, a webtool for the detection of clarithromycin resistance in Helicobacter pylori from whole-genome sequencing data
September 2020Link to article....
CCMetagen: comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data
April 2020Link to article....
Large scale automated phylogenomic analysis of bacterial isolates and the Evergreen Online platform
March 2020Link to article....
Improved Resistance Prediction in Mycobacterium tuberculosis by Better Handling of Insertions and Deletions, Premature Stop Codons, and Filtering of Non-informative Sites
October 2019Link to article....
LRE-Finder, a Web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in Enterococci from whole-genome sequences
March 2019Link to article....
Rapid and precise alignment of raw reads against redundant databases with KMA
September 2018Link to article....
Improved laboratory procedure and bioinformatics workflow for metagenomics-based identification of microorganisms: The workflow includes a curated reference genome database, reducing the identification of false-positives
April 2018Link to article....